Statistical methods for single-cell transcriptomics, epigenomics and translatomics
Our research develops innovative statistical methods and computational tools for genomics and transcriptomics, with a particular emphasis on single-cell data analysis. By combining statistical modeling, machine learning, and advanced computational techniques, we tackle the inherent challenges of high-dimensional, sparse, and noisy datasets to improve both accuracy and interpretability of analyses. A key focus area involves creating novel approaches for developmental single-cell RNA-seq data, enabling deeper insights into the dynamics of cell differentiation and lineage specification processes.
Relevant Publications
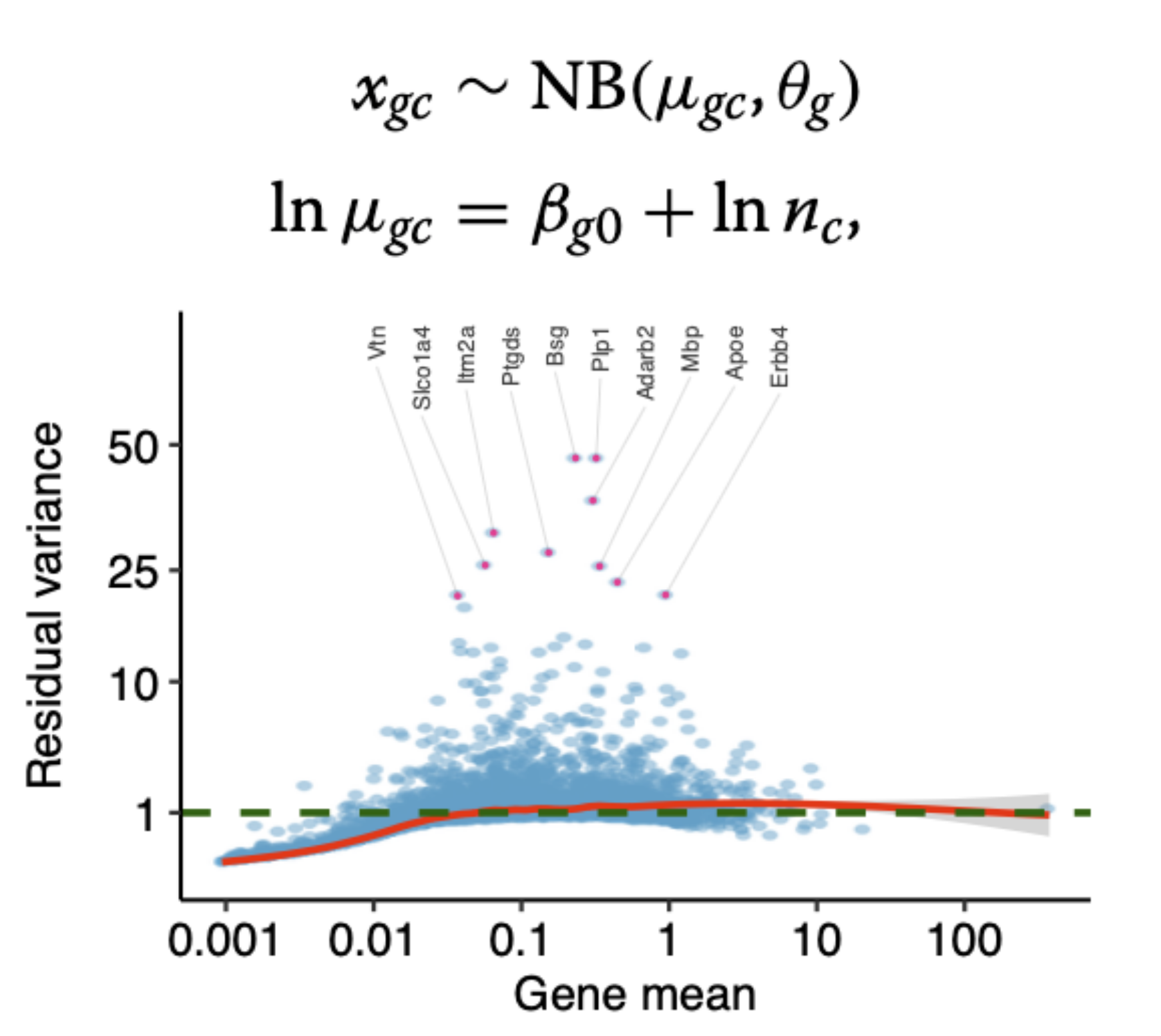
- Choudhary and Satija. Comparison and evaluation of statistical error models for scRNA-seq Genome Biology (2022)
- Hao et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nature Biotechnology
- Mundodi *, Choudhary* et al. Global translational landscape of the Candida albicans morphological transition G3 (2021)
- Choudhary, Li and Smith. Accurate detection of short and long active ORFs using Ribo-seq data Bioinformatics (2020)
- Choudhary. pysradb: A Python package to query next-generation sequencing metadata and data from NCBI Sequence Read Archive F1000Research (2019)
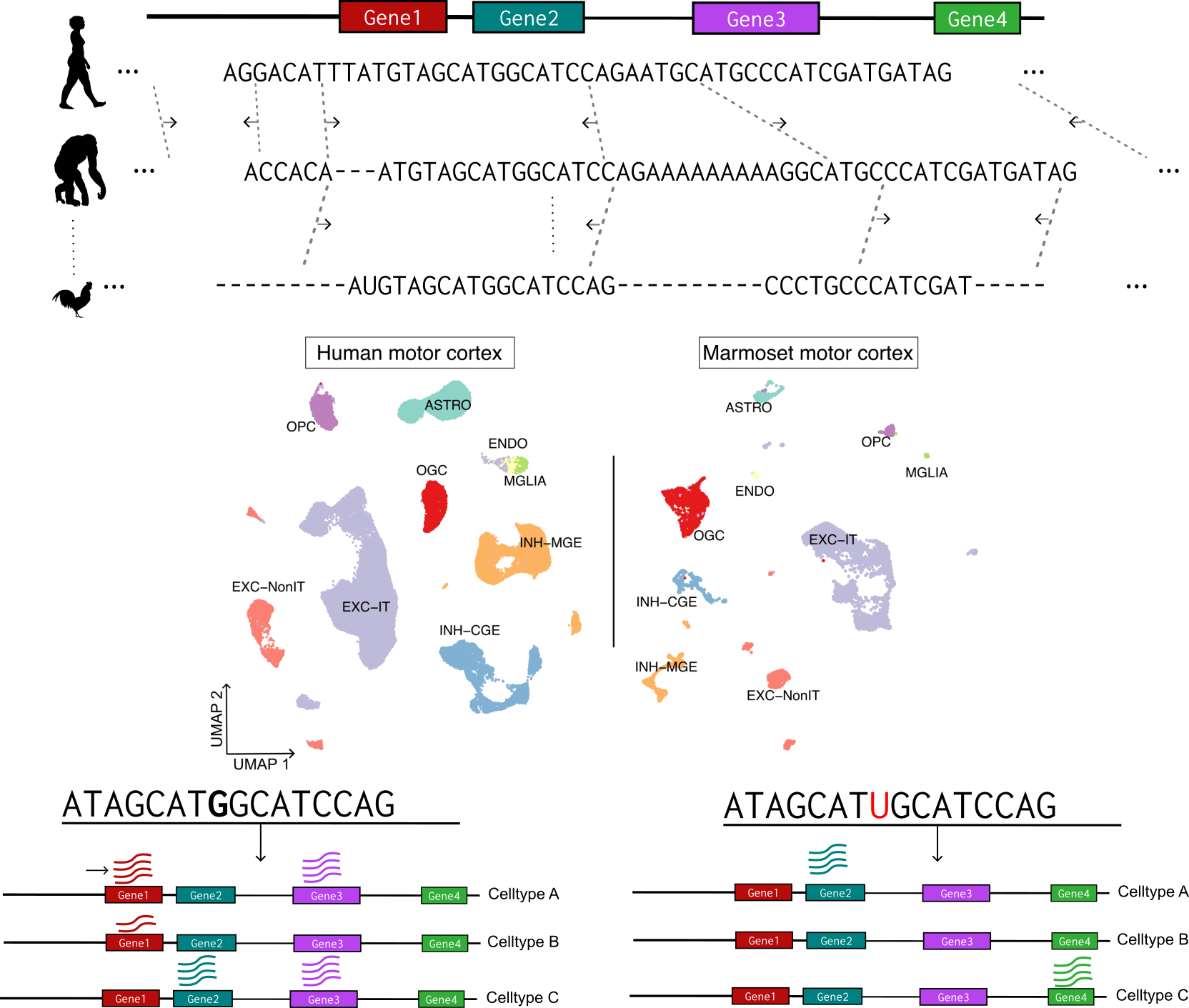
Integrative analysis of cross-species single-cell multi-omics data
A major goal in modern biology is deciphering the mechanistic principles underlying the phenotypic variation between species. The seminal works of Britten & Davidson and King & Wilson, more than 40 years ago, proposed that the phenotypic differences observed in species with high genetic similarity could be due to differences in the way genes are regulated. Deciphering how changes in the DNA sequence or the underlying epigenomic state have shaped the complex phenotype landscape across species remains a major challenge. We are currently using cross-species single-cell data to improve our understanding of human diseases and their underlying biological processes. Our current focus is on developing methods for cross-species integrative analysis of single-cell ATAC-seq and single-cell RNA-seq data ('multiome'). We aim to deepen our understanding of the non-coding genome by leveraging information across species thereby ultimately enabling us to accurately predict the functional impact of a novel non-coding mutation anywhere in the genome.
Statistical modeling and machine learning in public health
We apply statistical modeling and machine learning techniques to address public health challenges, particularly in the context of both communicable and non-communicable diseases. Our current research focuses on developing predictive models for risk analysis of breast cancer, and developing machine learning models for predicting obstructive sleep apnea. We are also interested in understanding the impact of environmental factors on health outcomes, particularly the impact of climate change on disease, birth and mortality patterns in India. These are relatively new areas for our lab, and we are excited to explore the potential of data-driven approaches to improve public health outcomes.
Current Funding
| Duration | Project Title | Funding Organization |
|---|---|---|
| 2024–2026 | Development of a Breast Cancer Risk Prediction Tool for the Indian Population | KCDH IITB - GKII JHU Breakthrough Research Grants Program |
| 2024–2027 | Characterizing non-invasive biomarkers of fatty liver disease using comparative single-cell multiomics | Anusandhan National Research Foundation (ANRF/ECRG/2024/006839/LS) |
| 2025–2028 | Development of machine learning models for predicting obstructive sleep apnea. | TiH (Department of Science and Technology), IIT Bombay |